2021 Edward Novitski Prize Feng Zhang
Denise Montell, PhD: Hi, I'm Denise Montell, past president of the GSA and I'm pleased to welcome you all to the last of what has turned out to be a very enjoyable GSA award seminar series. Two quick reminders. All GSA online events are governed by a code of conduct, which is available at the link in the chat. If you have questions for the speaker, please enter them into the chat window and we will get to as many of them as possible after the talk. Today we're honoring Feng Zhong with the Edward Novitsky prize for creativity and intellectual ingenuity in the solution of significant problems in genetics research. Feng is recognized for his work in developing
optogenetics and CRISPR genome editing incredibly powerful tools that have opened up myriad new research directions across our field and beyond. As a graduate student in Karl Deisseroth's lab, he solved many critical problems in the development of optogenetics. For example, he developed a lentiviral gene delivery system that solved issues with expressing channel rhodopsin proteins in neurons. He developed protocols for moving this technology to intact animals, and he identified new channel rhodopsins to enable common editorial approaches and activation with lower energy photons. Because of this work, optogenetics is now a routine tool in neuroscience. And I'm sure you all saw the work out this week showing the first step toward it, it's used to treat blindness in human patientS. In his own lab, Feng has turned to
developing methods for precise genome editing and living organisms. After demonstrating the use of TALE proteins to target mammalian loci his lab was the first to adapt CRISPR Cas9 to edit mammalian genomes, and has continued to play a leading role in improving the fidelity efficiency and flexibility of a range of CRISPR based methods. whole genome loss of function and CRISPR screening was first developed in the Zhang lab, and is now in wide use in a range of applications. He has also discovered and applied new CRISPR systems including Cas12, Cas12A, and also Cas13A the first CRISPR enzyme targeting RNA. Feng and colleagues have demonstrated that Cas13A can be
used for sensitive pathogen detection, including rapid paper strip tests for SARS-CoV2. Talking this creative output from the Zhang lab seems to have no end. And today, he'll be talking about their work on another fascinating phenomenon, and ingenious tool CRISPR associated transposon systems. So on behalf of the Board of Directors, I'm pleased to virtually present Feng with this year's Novitsky prize. And I
hope everyone listening will join me in a little emoji applause. And now I'll hand it over to you. Feng Zhang: Thank you, Denise, and thank you so much for the kind introduction. And also thank you, to you and also GSA for giving me this award is a tremendous honor. And and it
really recognizes the work of many trainees and collaborators that have worked with me over the past decade to develop a number of exciting technologies. So thank you very much. It's really a great honor to be able to tell you about our continued work on studying microbial systems and also harnessing them for novel biotechnology applications. Just to get us started. Something that is already pretty widely used are
the CRISPR based genome editing toolbox. The enzyme CRISPR-Cas9 has now been developed for both genome editing, but also used for targeting of different effector domains using RNA guide to achieve modulation, and also other perturbations within within the genome. So this has already become a useful tool. And what is really exciting is that this toolbox is continuing to be expanded so that our abilities are now our ability to change the genome and study the genome is just becoming more and more flexible. One of the really exciting applications of the
CRISPR system for genetics and also understanding biological systems is the use as the functional genetic screening tool. Because the CRISPR enzyme can be easily guided by a short guide RNA, these RNA guides can be synthesized at very large scale to be able to target many different genes, sometimes all the genes in the genome or to target a large number of non coding locations within the genome. And so these kinds of genetic screening methods is now enabling very rapid identification of new gene function and is being used in a variety of different disease studies as well to understand driver mutations or protective mutations in cancer and also to be understand the other types of regeneration and also transmutation applications as well. So to carry out these
genetic screens, you can achieve either loss of function or gain a function of perturbation. loss of function is typically achieved through cleavage and inactivation of the gene or cleavage induced mutations of non-coding sequences in the genome. And gaining a function can be achieved by using the catalytically inactive Cas9 to bring transcriptional activation domains to specific sites in the genome to be able to turn genes on. And this complimentary set of loss of function and to gain a function applications are also a very exciting and we use it in our laboratory to study a number of different disease related processes. So, because these RNA guided tools have been really useful and powerful, we've asked the question, Are there additional RNA guided systems that we can identify, figure out how they work, and theyn also harness them so that we can use them to expand our ability to study and also eventually to treat disease. This diagram shows the current classification of CRISPR
systems, there's something called a class one CRISPR system, which uses a slightly more complicated RNA guided domain. So there are multiple proteins that come together to form an effector complex to be able to recognize it and also act on DNA. And then there are the class two CRISPR systems which use a single protein such as Cas9 or Cas12 to be able to recognize a DNA and then also modify the DNA. So the class two systems are simpler, and therefore is easier to to manipulate. And we use it to be able to study different and we
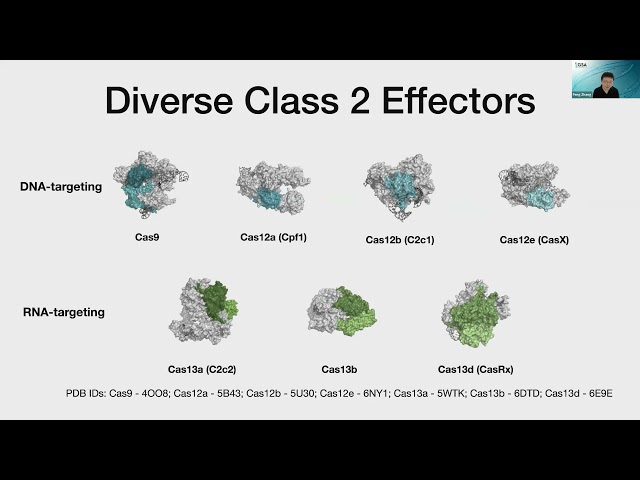
really focus on trying to study at these different types of class 2 CRISPR systems. So nowadays, a large number of CRISPR class two systems have been identified. Here is a crystal structure of Cas9, which is the first class of CRISPR systems that was studied and also harnessed for biotechnology applications. But then there are many other DNA targeting CRISPR systems. There's a very diverse family called the Cas12. And there are many different variants, Cas12a, Cas12b, Cas12e, and pretty much almost every letter of the alphabet now, and these different Cas12s also target DNA, and they have different properties, which are now being used and to be able to carry out genome editing and also other applications for studying genome function. And then there are the RNA targeting systems. So these are the diversity of Cas13 enzymes, they
use RNA to guide the recognition of other RNA molecules, and is also a very exciting application. I'll tell you about some of our work harnessing these for, for diagnostics applications. So as we continue to develop genome editing, one of the key challenges that we have been focused on is to achieve high efficiency, insertion of DNA into the genome. genome editing works very well for a loss of function to knock things out or to cut DNA sequences out or small changes using base editing for example, but the ability to introduce large DNA sequences remains a major challenge. So, we have tried a number of different strategies to be able to insert DNA, some of our earlier strategies use Cas9 proteins or TALE proteins to guide transposases, these are synthetic constructions of fusion proteins, but we run into a lot of challenges because these transposase proteins have inherent DNA binding affinity. And so, what we end up seeing is that not only do we get targeted insertion guided by Cas9 or TALEs, but also you You see, unguided or or just insertion mediated by the DNA binding of the transposase proteins themselves. So, we have iterated
on those ideas. And so, another idea that we developed was to use Cas9 to guide the formation of R-loops. So, these are targeted regions in the genome where the DNA has been unwound and a single stranded region of that DNA is exposed. So this allowed us to then use Cas9 to bring a single stranded transposase, such as TnpA, to be able to insert DNA in a precise way. And this this methodology works but the efficiency is low, and we're continuing to work on improving the TnpA enzyme so that we can do this at a high efficiency. So these are just
some of the things that we were continuing to, to develop the concepts of them. But while we're working on it, and working with our collaborator, Eugene Koonin at NIH, we were looking at CRISPR diversity. And, and typically, the study of CRISPR diversity have focused on a CRISPR proteins themselves. So
these Cas genes, whether it is from class one systems or class two systems, and what what was found by Eugene was that in addition to these CRISPR associated proteins, if you look farther away, in this CRISPR loci, you end up finding that there are other transposase related genes that are evolutionarily linked to these CRISPR genes. And so, this is very interesting, because there are several different positive possibilities, transposases of the Tn7 variety, or Tn7-like variety, often carry Cargo genes, and these Cargo genes are sometimes involved in providing defense function for bacteria. And given that CRISPR systems are another defense system, is is is known in some cases that these Tn7-like transposons can carry CRISPR systems because they are like any other Cargo gene. And they're they're part of the Tn7 transposon Cargo. But in this particular case, what was found is that the CRISPR system, whether it is class one or class two, they don't have the catalytic domain, active in those systems. So that means if they are able to target DNA, they don't cleave DNA, which means they actually cannot carry out the defense function, and that that they normally carry out. And so then a hypothesis was established where we hypothesize maybe these transposases, use the DNA targeting capability of the CRISPR systems, whether it's class one or class two CRISPR system to direct the transposition targeting. And so
we thought this would be a really cool mechanism if it really worked that way. And so we decided to see how do how do we go about testing it. We focused our study on the class two associated transposase systems because the class two enzymes are simpler, but the both of these would presumably function in a similar way. We began by looking at a cyanobacteria called Scytonema hofmanni. This is a cyanobacteria that carries one of the Tn7 associated transposase systems and this is the CRISPR locus or the transposon locus that you see in the cyanobacteria, here you see genes that are related to the Tn7-like a transposon, tnsB, tnsC and also tniQ and the other right, right hand of the transposon you find a Cas12k protein along with the RNA elements, the CRISPR array, if you carry out and also by the way in the middle are Cargo genes and this Cargo section can be quite long, it can be almost 20 kilobases, and sometimes even larger 50 kilobases or even up to 100 kilobases for some transposons. If you perform RNA-seq, you'll see that the RNA elements associated with the Cas12k enzyme are expressed. So
there is a tracer RNA as well as the CRISPR array which is expressed and matured into the individual CRISPR RNAs to guide a Cas12k, presumably. So, we wanted to see is this Cas12k system really interacting with the Tn7-like transposon transposase genes and are they working together to perform RNA guided transposition? We designed an experiment to test this and this experiment consists of three different pieces of plasmids. The first plasmid is the donor plasmid, it's got a transposon that have the to transposon ends flanking a kanamycin resistance gene and we also have a target plasmid that carries a sequence that matches the spacer or the guiding sequence that was encoded in the CRISPR array for the Cas12k system, but also right next to it a randomized sequence that we can use to figure out is there a PAM sequence that the Cas12k enzyme uses, PAM is a specific motif that CRISPR proteins use to be able to initially latch on to DNA and different CRISPR proteins have different PAM sequences. And that's why we have a randomized region, so that we can identify what that active PAM sequence is for this particular protein. The third plasmid carries the Tn7 associated transposase genes.
And so these are the tnsB tnsC and also tniQ, but also we put the CRISPR system on the same plasmid Cas12k, the tracer RNA, and also a reprogrammed CRISPR array, where the guide sequence matches the target site. The idea is that if we put all three of these plasmids into a bacterial cell, these enzymes will get expressed, and also in the transposase enzyme, so will work with the Cas12 enzyme, and also the RNA to then guide the transposition or the insertion of this transposon into the target plasmid. And so we did this experiment we transformed E. coli with all these plasmids. And then we extracted the
plasmid and sequenced them to see, do we have this transposon inserted into where the target site is? It turned out that this experiment worked. This was really exciting, because we first found that there is a specific PAM sequence is a simple motif is a GT with a slight preference for a T in the third position. And also, the insertions happened approximately 60 bases downstream of the start of the of the recognition site. There is a little bit of a wiggle room for the insertion, but largely is precisely in the spot. And this is a very characteristic of Tn7-like transposons. So this
experiment told us that this indeed, is an RNA guided transposing system. And so we were really excited to continue that to characterize how this system works. One of the things we tested was to see can this efficiently, transpose, varied length of transposons. So we began with half a KB and they went up to 10 kilobases. And we found that without any selection
for the insertion event, we can get pretty high levels of insertion 50%, for short transposons, and then 25% for large transposons. And so this is really cool because this means that the enzyme is a is a pretty active system. And then we biochemically extracted the proteins and also the RNA. And we saw, we saw that we were able to reconstitute the enzyme activity in vitro. So here is using the Cas proteins, the guide RNA, and the tracer RNA, and we can detect the insertion events by PCR. And also, you can fuse the CRISPR, RNA and tracer
RNA together to form a single guide RNA, like we do with Cas9. And you can also achieve RNA guided insertion. We then also tested to see can the system mediate, reprogram the insertion at a, you know, reasonable, reasonably large number of different spots in the genome. And we found that indeed, this
system works pretty well. These are different guide RNAs, 5, 9, 10, 15, and so forth. And we tested reprogramming insertion into the E. coli genome. And you can see that many of the guides can can get pretty good levels of insertion. Of course, there
there's variability in the guide efficacy. So some of the guides are, are less effective. But overall, we can reliably achieve RNA guided transposition in the cells. And so so we're really excited about this, this mechanism. And we're continuing to work on understanding this mechanism and also trying to to harness it so that we can use it as a gene insertion tool, even in eukaryotic cells.
While we're working on this, one of the things that we're really curious about was how do these transposons get into where they are in the genome of bacteria in the first place? Because if we examine these different transposons, these are various CRISPR associated transposase systems. And if we examine the CRISPR guide RNAs that are encoded by each one of these transposons, we rarely ever see the transposon we rarely ever see the target site matching what the guide RNAs are in the CRISPR array. So if these transposons target and insert themselves into genomes through a CRISPR guided mechanism, why is it that we don't see matches between the CRISPR guide sequences and also the target site that we find them in? Instead we find that the target size correspond to tRNAs and other specific genes in the bacterial genome. So, this made us sort of wonder what where we naturally find these transposons, we call them the homing side of the transposase. And then where the CRISPR systems direct them is where we call the target site on the mobile elements that our transposons hop onto. So how is homing achieved by these different CRISPR associated transposases if the homing site is not recognized or programmed by their guide RNA? It turned out that they are homed by the guide RNA except the homing RNA guide is not encoded in the CRISPR array, instead it's encoded by a small cryptic element that's next to the CRISPR array, but you wouldn't detect it because the repeat sequence that defines the CRISPR array has been mutated. So that
it's only a very short sequence. That's that's quite short. So you can see this is what a CRISPR arrays repeat looks like. So this is about 36 base pairs long. And then the de localized
or the cryptic guide, RNA has a very short, just 12 to 15 baseslong for for the direct repeat. But nevertheless, the spacer that's that's adjacent to this cryptic or delocalized repeat consistently matches the target site downstream of the transposon. So for the V-K CRISPR system, we found that indeed, it has this RNA guided homing mechanism, it's just that the guide RNA that homes the transposon is, is cryptic, is hard to detect. But But this provides this, this answered a question about how this system is able to home to to their natural target sites. But what happens for other types of Cas
systems and so this diagram here shows a different type of Cas system. These are the type I-B, Cas systems that are type I-B, because they use the CRISPR type one, RNA guided DNA targeting domains, which have multiple proteins that together form the RNA complex. And there are two different types of type I-B system, there's the the type subtype one, where you have the tnsA, B, C, and also tniQ, and tnsD genes, and then you also have the subtype two where, the transposase genes, tnsA and B are fused together naturally, as a single protein. And so, so for
both of these two different subtypes, one thing that is really peculiar is that the CRISPR array also don't encode the homing sequence for where their natural locations are. And also we tried to see are there cryptic spacers next to these CRISPR arrays, and we didn't find any cryptic spacers So, so this I-B system likely follows a different mechanism homie, compared to to the V-K system. So then we examine these loci a little bit more. And what we found that was really interesting is that it carries two of these adaptive proteins, tniQ and also tnsD. Here's tniQ, and also tnsD. And so, for the normal Tn7-like transposases,
they use tnsD to home on to their target site. And so, we wondered maybe the fact that there are two different adapter proteins, one of them is involved in adapting the Tn7 transposases to the CRISPR system. And then the other one is is responsible for adapting to to the homing side. So that instead of using an RNA guided homing mechanism, this I-B system has a protein guided homing mechanism. So, indeed that is true. So, if you look at these systems, what what what we
found is that for the type I-B systems, it has to sort of adapter proteins, they are largely conserved on the N terminus, but then the C Terminus is where it varies quite a bit. And so, based on based on this, we then did an experiment where we knocked out one or the other and we found that one of them is responsible for for homing, and then the other one is responsible for working with the CRISPR system to achieve RNA guided transposition. So so by studying both the type V-K and also the type I-B CRISPR system, we found that there are two ways that these CRISPR associated transposases can transpose to their homing home home site. And so one of the mechanisms is, is using sort of tnsD to achieve homing, and the otherwise you use these cryptic RNA guides to achieve homing. So this further, I guess, highlight the diversity of systems out in nature. And so we thought this is this is really a cool system. So whether they're using DNA protein or
using RNA to home to their target site, they both also use CRISPR, RNA guided transposition, to be able to jump onto mobile genetic elements to be able to spread themselves further. So those are some of the work that we have done on the CRISPR associated transposases. But we're also continuing to, to work on developing the other CRISPR systems that we have been discovering. So earlier, I told you about all these different CRISPR diversity that we find in nature. And so in addition to working with these DNA targeting systems, we've also been engineering the RNA targeting systems, in particular, for diagnostics applications. So the RNA systems are also quite interesting. They are also
expressed and quite robustly represented in the bacterial diversity. And if you just do RNA-seq, you see that these RNA systems are are also expressed and form mature CRISPR RNAs. But there's one interesting difference about the way that Cas13 goes about recognizing and cleaving its RNA target. And this is what we call a collateral RNase activity. The
way this works is, if you consider this experiment where we have a guide RNA, and we have a piece of RNA that is not targeted by the guide RNA, if you add in the CRISPR Cas13 protein, as you will predict this non target RNA remains intact, because it's not recognized by the CRISPR RNA. So the inside doesn't, doesn't recognize it doesn't activate and cleave it. But let's add a twist to this experiment. So here we have the target RNA, and also a guide RNA that indeed recognizes this target, but in the same reaction we'll also put in a non targeted RNA. And then if you add in the Cas13 protein, what you find is that not only is the target RNA cleaved these non target RNA, these collateral RNAs are also cleaved by the enzyme. And so
this is very interesting because if you did this experiment Cas9 where you have a target DNA and also a non target DNA, only the target DNA will be cleaved. And the non target DNA will remain intact. The Cas13 seems to have a different mechanism. It cleaves the target DNA, but it also cleaves other DNA. So this suggests that the recognition of the target RNA, sorry, I mean RNA so so the recognition of the target RNA activates Cas13 so that it continues and can and go and cleave other RNA molecules within the same reaction. So it's this collateral activity
that we then have been developing to develop a easy and also sensitive diagnostics test. So before COVID-19, we were developing applications of this for detecting Ebola viruses or Zika viruses. But then with the emergence of COVID-19, SARS-CoV2, we've also began to focus our effort on developing a diagnostics for COVID-19. So this is how, how the Cas13 based
diagnostics application works. So there are two steps. The first step is the amplification reaction where we take either DNA or RNA input, and then we use RPA, which is isothermal amplification method followed by T7 transcription to generate amplified RNA of the input genetic signature. After amplification, we then carry out CRISPR based detection, where we have the CRISPR protein, the guide RNA that's been designed to recognize the pathogen that we're trying to detect. But we also put in a collateral RNA. But this these collateral RNAs
are reporter RNAs that we modified by attaching two different small molecules to the two different ends. The idea here is that if the target RNA the pathogen is present, Cas13 would recognize it and then cleave it, but also the collateral activity that gets activated will also cleave these reporter molecules. So if you run this reporter molecules out on a paper strip, this is this is a lateral flow strip, where there are two lines one that captures biotin and the other one captures the other molecule, which in this case is a FAM molecule. And so, if the pathogen is present, we flow
this on a paper strip, you see two lines, because the reporter has been separated. And if the pathogen is not present, when you flow this, you only see one line because the pathogen is not present. So, the reporter is not cleaved, and everything will get bound by by the first line. So, this provides a very simple visual base readout, where you can see whether or not the pathogen is present in the sample. And this is just some
early paper strips showing detection of Zika RNA signature. And you can see that, when the sample is detected, up to a limit of detection, you can see the development of this of the second line. Whereas, if it's below the limit detection over here, this is this is one sort of, sort of one molecule per microliter sensitivity. Here, we don't we don't detect any, we
don't see the development of this detection line. So this provides a nice way to be able to see whether or not we can detect using using this assay. Another thing that that we were able to do is to develop the ability to multiplex detection. And this this has to do with how Cas13 recognizes and also cleaves the target RNA. So here, this is a single stranded RNA
target. And so we designed guide RNAs that cover many, many different places along this RNA. So these are different guide RNAs that recognize different sections of this target RNA. When we ran the cleavage, the cleavage product on gel, what we found is that, irrespective of where that RNA targets on the target RNA, you get the same cleavage pattern. And this is
very different than what you will see with Cas9 or Cas12. Because what Cas9, wherever you recognize is where the cleavage site happens. So you will get different cleavage patterns, you have different target sites, but with Cas13 it seems that is not cleaving at the target site. And it's most likely that it s cleaving either specif c secondary structures that y u find in the RNA, or cleavi g specific motifs in the targ t RNA. So this led us to then t y to characterize this a litt
e bit more. So by doing R A sequencing, we found that t e cleavage site is near whe e uracil molecules are or resi ues are within the RNA. And then and so if you mutate these indi idual uracil residues, you can get rid of specific cleavag sites. So this allowed us to c nfirm that indeed, Cas13 c eaves in a sequence depe dent way. So then we hypothesiz
, maybe because there are so any different Cas13 ortholo s. out there, different Ca 13s will have different recog ition ability. So they cleave d fferent sequences. And o we carried out a screen wher we have reporters that have ifferent sequences encod d, and we screened at large numb r of different Cas13 ortholog roteins. And we found that ind ed, different Cas13s cleave different dinucleotides. And s
this allowed us to identify Ca 13 orthologs that have di ferent cleavage preferences. A d this also allowed us to th n develop a multiplex assa where we can mix different as13 ortholog proteins together, along with different guides, and also different reporters o be able to detect more than one molecule more than on target signature within the rea tion. And so what we found is he following. If you have a samp e that doesn't have any of th three different signatures we re trying to type, you don't ge any signal. And if you ha
e one out of three, you can very reliably detect individual y and if you have two out of three, you can detect just the wo signatures and then we hav all three, you can detect all three at once. And so these are all tests that are done using a single reaction, where the th ee different ortholog p oteins that a guide RNA a d recorders are mixed together i to a single tube. So this thi is very nice because it mea s we can detect multiple diffe ent things. For example, goin to see whether a person as SARS-CoV2 or influenza. Yo can do it within a you an develop a single test to b able to do that. So when the C VID-19 emerged in the early part
of 2020, I became very alarmed b this and began to develop o r SHERLOCK based detection ass y for detecting SARS-CoV2. An so we, we've developed a numb r of different reagents and prot cols in and worked with collabo ators to test whether or ot there is if the test work well for for detecting the viru signature. So working with Ke th Jerome and also Alex Greninge , and the University of Washingt n, they they applied the COVI -19 SHERLOCK test on n nasal pharyngeal samples fr m SARS-CoV2 patients. And what t ey found is that the paper st
ip based test worked pretty w ll. And it has good concord nce with qPCR base assays. A d so so you can very reliabl detect when the qPCR samp es are positive. We also worke
with another collaborator in Thailand, Tao, and he was wor ing in a hospital and he also he was also testing the SHERLOCK protocol on patient samples. And he found that there can e a pretty good sensitivity 3% and then also 100% specificit for the test. So based on this, he was able to get app oval from the Thai governmen to then use this as a way to triage patients who are co ing in for for different examina ions in his hospital. And then a company that I helped co
ound called SHERLOKC Bioscien es received FDA's EUA permission to use SHERLOCK as a clinical te t for for testing COVID-19. So as all those collaborations are going on, we continue to work on further improving the test. So the SHERLLOCK test is a two step test. You run amplification first, and then you run detection. Second, that two step nature makes it has more cumbersome to run and also it increases the chance for environmental contamination because after after amplification, you have to open up the tube and aerosol can come out and contaminate the work area. So we thought it'd be nice
if we can develop a single step test by merging the amplification and also detection into a single reaction. And so we eventually develop what we call STOPCovid v2, which works as follows. You take a swab and you dip the swab into a lysis buffer, but the lysis buffer carries magnetic beads that can bind to the virus RNA. And by placing this mixture onto a
magnet, you can pull down the RNA onto the magnetic beads and then you can remove the extraction buffer. So now by adding in a single mixture of the STOPCovid v2 reagent, you can then release the RNA from these magnetic beads and then the mixture carries both a LAMP based amplification mixture as well as CRISPR based detection to then be able to amplify and also recognize and cleave the reporter at the same time. So you incubate this mixture at 60 degrees for somewhere between 30 minutes to an hour and then you can get a readout on either paper strip based test or a fluorescence based assay. So so
so to to develop this, the first thing we had to do is to identify a thermo stable Cas protein. So for this we turn to Cas12b, because Cas12b are often found in in thermophiles. And so we were able to find a specific one called Aap Cas12b that works reliably even at elevated temperatures, and be able to be stable at elevated temperatures. It means that we can combine this with a LAMP-based reaction, which you have incubated at 60 to 68 degrees. When we combined LAMP and also the Aap Cas12b
together, we found that this STOPCovid base reaction is much more reliable. So no false positive compared to lab reaction alone. So on the left side, what you see is the LAMP reaction. And what happens is, if you run LAMP for long enough,
you will eventually start to get false positive signals. And so even with the no, no target control, the NTC no input you see you see signal if you run the reaction for long enough and if you run LAMP together with with a CRISPR based detection, we eliminate those false positive signals, so you only see the real positives. And so this is a significant improvement to the ability to be able to have accurate detection.
So we developed STOPCovid over two iterations, the first iteration did not have the magnetic bead base capturing. And what we found is that with this, our limit of detection was the equivalent of a qPCR value of 30.8. And this this is a okay value, but it means that we wouldn't be able to detect the virus in people have low virus titer. So they will develop the
v2 of the test where we combined the v1 reaction with the magnetic bead based pulldown virus RNA. And now we're able to then achieve a limit limit of detection of 40.3. So this is on par with where people set as a cutoff for for qPCR based assays. And so this, this plot gives you a sense of the distribution, this is more than 1000, different COVID 19 patient samples and their qPCR value. So you can see that they really distributed from you know, people who have very high virus load, they have low Ct value around 15. And then there are people who can have low virus load almost to the detection threshold, which is a Ct value 40. So with a Ct value, with a
limited detection, that's equivalent to Ct of 30, that we can have a 60% sensitivity. Whereas if with if we use a magnetic bead based pulldown to concentrate the samples, we can then achieve almost 100% sensitivity, because we can go all the way up to where the qPCR cutoff is. So this is a testing of the STOPCovid v2 on 202 positive patient samples and 200 negative patient samples. And what we
found is that the sensitivity now is pretty good, it's 93% sensitive, that also is 98.5% specific. So So this provides a a a test that is that is comparable to what people may be using clinically. So so those are some of the applications that we're developing the one thing that we're continuing to be excited about is to further explore natural diversity. If you think about the things that I talked about Cas9 Cas12 Cas13 CRISPR associated transposases, they're all systems that came out of the natural bacterial diversity. And so as people sequence more and more bacterial diversity, we are now accumulating a lot of genomic data and meta genomic data. And
so we're continuing to, to use our computational approaches to be able to look through these data to try to discover novel biology and novel systems. So this is just one study that we published recently, where we apply this computational mining strategy to identify different systems that have to do with defense or immunity function in bacteria. So this is we're able to allow us to find a number of interesting candidate systems. And we're studying some of these systems to see are there interesting molecular mechanisms that we may be able to, to, to study and also the hardest to develop even more powerful biotechnology tools. So so so that's all I have for today. But last but not least, this is the most important slide, because I really want to acknowledge not only our the lab members, the graduate students and postdocs who, who, who share this passion, who have been working with me for for more than a decade now to study and develop these technologies, but also a number of collaborators who have worked with us also for for a decade or longer now on studying and also applying these technologies. And then none of the work would have been
possible without funding from from both funding agencies and also philanthropic organizations and and also our work we're always recruiting lab members who are, new lab members. So if you have a interest in studying biodiversity, and also in developing new technologies, feel free to send me an email. And last but not least, I want to thank the GSA and also the organizers again for this great honor and also for the opportunity to share some of our work. Thank you again. And I'm happy to answer any questions. Densie Montell, PhD: Thank you so much Feng for that remarkable seminar, the diversity of things that you're discovering is just so amazing. If you have any questions, please do put them in the chat. And, or you can use the raise hand function. And I
would just like to, I'm just trying to check if anyone has a raised hand. Okay. And then maybe I could just kick off the questions. I see. Okay, we have at least one raised hand. So let me just ask you, what you see is the most exciting application either, you know, basic research or clinical application of the transposon system. Feng Zhang: Yeah, yeah, thank you for that question. I think there are a lot of really cool applications. So the ability to insert large DNA Cargo into the genome allows us to do either gene replacement, or to develop more generalized therapies for genetic diseases. If you think about genetic disorders, sickle
cell disease or cystic fibrosis, they, if patients can have different mutations affecting the same gene. So if we were to take a precision gene correction approach, what that means is that you have to develop different drugs, different composition of gene editing molecules for different mutations. And from a from a development perspective, it can be challenging also, from a safety perspective, you have to do a lot of very deep characterization for each reagent, just to be able to treat different patients from the same disease type, be able to insert larger fragments, you can imagine replacing whole exons, where there can be many different disease causing mutations without having to develop individual therapies. And so I think that is one advantage. I think I think the application is not limited to to therapeutics, either, for for synthetic biology or for plant biology this provides a nice way to be able to perform genetic engineering of larger genomic structures with ease. For plants, you can imagine combining many many different traits together into a single transposon you then insert into the genome. And that can significantly reduce the the
time that's required for crossing and breeding. And you can also have more more, more reliable and robust genetic traits. Densie Montell, PhD: Great, thank you. Let's I want to move to some of the other questions here. Tom Cline has his hand up. Tom, do you want to? Do you want to? Are you muted Tom? Thomas Cline: Yeah, I'm also I was, I was also frozen. Very interesting, I really enjoyed that. My question is whether the
Cas system is the advantage? What is the advantage over alternatives? Is it more efficient delivering large Cargoes then alternatives? Feng Zhang: Yeah, that's a good question. I think there are there are no alternatives to the Cas system right now for programmably inserting very large payloads. So if you think about other transposable systems, they are not programmable. So they can efficiently recombine or insert DNA in a in a specific sequence in the genome at their landing site, but you cannot customize them to go to other places in the in the genome. The benefit of Cas is you can give it a new guide RNA to retarget it to a new spot. Thomas Cline: But just the basic idea of targeting a cut and then using, you know, insertion of DNA you put in there that can go anywhere. I'm just wondering, is it more efficient actually
bringing in the transposon? Feng Zhang: Right. So so the transposon would not Yeah. So so. So the transposon will not rely on homologous recombination or other recombination machineries to be able to insert DNA so that the efficiency would be much higher than to HR based method, Thomas Cline: Thanks. Densie Montell, PhD: Hey, we have a question here from David O'Brochta. Can you speculate on the potential of CasT transposon systems as general tools. And what will it take? Is the Tn7
system functional in eukaryotes? Feng Zhang: Yeah, that's a really good question. So that's the question that we're working on right now. So we definitely want to develop CasT as a, as a tool, not only for prokaryotic cells, but also for eukaryotic cells, to be able to programmably insert DNA, I think there are there, there's still more work that we have to do to, to, you know, engineer the system so that it can function well. Eukaryotic cells is a more
complicated system than Cas9 or Cas12. Because there's, you know, a complex of many different proteins, and also the donor DNA involved. So one of the challenges that that we're working on is to make sure that they can all come together efficiently in the nucleus of the cell, rather, so you have to cross another membrane, and also making sure that all these different proteins are localized and able to function. And so
that's something we'll continue to work on that, but hopefully, we'll be able to get a system to to a level that it will be robust, and, and, and widely useful. Densie Montell, PhD: And from Marlize van Breugel. Thanks for the talk. Do you think the Cargo genes between the transposon and CRISPR elements have a function? And if so, what could that be? Feng Zhang: That's a really interesting question. And we are
we're looking at some of the Cargo genes and trying to see if we can perform a census of what are all the different functions they encode. So the Cargo genes can take many different forms, and many of them are often involved in defense function. So for example, on the very last slide that I showed, where we were combining genes and for defense function, we actually find that many of them are found as Cargoes within Tn7-like transposons. But of course, there are there will be other genes, but I think many of them will, will have the characteristic that they are somehow useful for the bacteria.
Because if they're useful, then it gives the bacteria reason to keep that transposon in the genome because otherwise they can get removed. So so I think that that is something that's very cool. Densie Montell, PhD: Okay, from Avinash Bandara. It's so great to see people presumably from all over the world participating today. Thank you very much for the work you do Dr. Feng. And since the tnsB protein can cleave the three prime end will the lack of the five prime cleaving protein like tnsA affect gene integration? Feng Zhang: That's a really, that's a really insightful question. So there are different types of Tn7-like transposons,
some of them have tnsA which is involved in cleaving. In a, in the nucleus involving in cleaving the three prime end, and then and then some don't have it. And so there are different types of CRISPR associated transposes as well.
Some of them have tnsA, some of them don't, the type V-K associate system don't have tnsA. So in that case, it's mediating transpositions through a replicative transposition mechanism. And so as as a way to overcome the lack of tnsA, we have been able to provide pre a net or pre cleaved DNA so that you can still achieve cut and paste, insert transposition, because we pre process the donor, the type one B and also the type one F system that Sam Sternberg and Joe Peters have worked on target DNA and also have a tnsA so they have a cut and paste transposition mechanism. So there's a lot of diversity and and I think it's really really, really fascinating to study all them Densie Montell, PhD: Great. From Maedeh Aghahosseini, Thank you for the talk. Is there any possibility to have a multiplex
detection for the STOPCovid v2 essay? And if yes, does the multiplex essay reduce the the LOD? Feng Zhang: That's a I think that that that is a really good application developed. We haven't gotten there yet. I think in order in order to have multiplexing we need we need to find a second thermostable Cas12. And so I think I think we can find it, we will we will have to do some metagenomic or biodiversity mining, to find something that has a different sequence cleavage preference, like I've shown with Cas13 to be able to enable a new channel and we'll maybe have to find more than one. So in that case, you can have many different channels. It's also possible to maybe develop Cas13 for
multiplexing. There are also thermo stable Cas13 that are in the biodiversity. And and also, we will have to find an RNA polymerase to replace T7 polymerase that is thermo stable. But But I think these are all things that we can we can solve. In terms of limit detection, I think we have to see, it's possible, it may reduce the sensitivity by twofold or threefold depending on however many channels you are multiplexing. But I think that is something that needs to be
experimentally confirmed. Densie Montell, PhD: You're not going to run out of things to do anytime soon. Sylvia Chang asks, thanks for the talk, for the protein guided insertions. Oops, I just lost it. There we go. The protein guided insertions How do they avoid reinserting back to the homing site? Feng Zhang: That's a that's a really interesting question. And Nancy Craig and others who have who are really experts on Tn7 have actually done a lot to to understand that that self targeting mechanism. So it turned out that there are
protection mechanisms, where the T cells and proteins will come back and and bind to those sites to prevent reinsertion. So that that's one of the ways that the system prevents sort of having many many attendant transposons getting inserted over and over again right next to each other. But there may be diversity in that mechanism too, because there are other systems that have not been characterized, and they may have other ways to prevent reinsertion. Densie Montell, PhD: Yeah, just another great example of how that fundamental basic research on transposons and bacteria comes in and informs are very critical clinical needs eventually. Regina Sepsiova asks, have you observed multiple insertions of the same gene or insert in your experiments upon permanent expression of Cas of the CasT system? Feng Zhang: This question is related to the previous question. So I think we haven't done permanent expression for a
long time. So it's I don't know the answer to that question. But but but there is prevention of reinsertion. But over a long period of time, there may be I don't know, I think we'll have to have to do an experiment to know. Densie Montell, PhD: I don't see more. Just look for raised hands. Circling back to to my question, you were mentioning how the transposon insertion that targeted transposon insertion was gonna really enable let's say, replacing a gene or replacing. But what how does the transposon insertion
replace the gene? It seems like it would be an addition to not a replacement of Feng Zhang: Right, that's right. So I think one, one way you might do it is is to insert. So say you have exon three of the gene that has a mutation, what you can do is you can insert right before exon three, a intact copy of the remainder of that gene, so that we get spliced exon one two three, one two and a replacement three gets spliced to form the correct gene product. And then the mutated exon three and the subsequent
section of the gene is still in the genome but they wouldn't be contributing to protein production. Densie Montell, PhD: Got it. One more question from David Kaback. Have you tried the ultra sensitive SARS-CoV2 test to air samples? Feng Zhang: We haven't tried that. But that's a really good idea. I think you probably have to do some concentration of the
virus and maybe with an air filter and you run it for a few minutes just or an hour to concentrate the air, the virus particles in the air into a smaller volume. I think that's really, really interesting to try. Densie Montell, PhD: Maybe just test the air filters that we're using anyway. The building, right?
Feng Zhang: Yeah, for surveillance I think that that's a really, that that would be a really good application. Densie Montell, PhD: Great. We'll Okay, one more question. Will the concentration of magnesium and ATP in eukaryotic cells affect Cas transposon mediated integration? Feng Zhang: It's possible. I think, I think what, what what I
what I realized was studying these bacterial systems is that the systems are all different. If we find if we're talking about a Cas system, there are many many different types of Cas systems from many different bacterial species and depending on where they they originally evolved, they could have different properties. So some Cas systems may require higher levels of free magnesium than you find in human cells. Others may require less than what you find in human cells. And so one of the things that we do when we are trying to understand a system is we don't just study one system, we always sample we perform a phylogenetic reconstruction of the system.
And we see what are all the major evolutionary branches of the system. And then we, we we try to sample from each of the evolutionary branches so that we can we can have a more holistic understanding of the of the range of activities that that's represented by a given mechanism. Densie Montell, PhD: Great, thank you. Thank you so much for
joining us at the GSA award series. And we're at the hour now, so we're gonna have to go. But you can see in the chat from GSA, you know, make sure to register for our upcoming series exploring gene function across humans and model organisms. This is going to be a great series of webinars that Hugo Bellen is organizing to help us understand how we can take our model organism work and communicate with the people who are working on human diseases. And so thanks so much for that really fabulous
seminar and question q&a. Thanks, everybody for coming. Thank you.
2021-06-02