SIP technologies at EMSL and JGI Webinar
REX MALMSTROM: All right, well, I'm going to get started here. Thanks for joining us today. I'm Rex Malmstrom. I'm a scientist at the Joint Genome Institute.
And today, we're going to discuss the various capabilities that utilize stable isotopes and that are offered by the JGI and the Environmental Molecular Sciences Lab. It's our hope that this webinar will provide a better understanding of how these technologies work and how you might access them in order to advance your own research. Please feel free to send your questions to us through the Q&A feature in Zoom, and we'll take some time at the end of the presentations to answer these questions. Before that though, just a little bit about us.
The Joint Genome Institute is a US Department of Energy user facility and we're located in the Lawrence Berkeley National Lab. We're world leaders in the genomics of microbes, plants, fungi, algae, and complex environmental communities. More specifically, we use our expertise in the areas of DNA and RNA sequencing-- the DNA synthesis, metabolomics, data analysis-- to help researchers characterize how these organisms carry out complex biogeochemical processes. And we also explore how they can be harnessed for sustainable biofuels. The Environmental Molecular Science Lab or EMSL is also a DOE user facility and they're located in the Pacific Northwest National Lab. EMSL has expertise in a wide variety of areas, including imaging, proteomics, chemical analysis, and modeling.
In particular, EMSL has a wide variety of methods focused on stable isotopes and how to trace the flow of elements and energy through plants and microbes in soils and many different things. And we're going to hear more about that today. Scientists like yourself can access the capabilities of JGI or EMSL through different calls for proposals that are offered throughout the year. But there is an opportunity called the EMSL-JGI FICUS that allows users to apply to both facilities in a single proposal. And there's a particularly nice synergy here between the two user facilities around stable isotopes, and that's the focus of today's talk.
You can learn more about FICUS program through the two websites on the slide here. However, the FICUS letters of intent-- the deadline for submitting a letter of intent for FICUS, it just passed. So if you didn't submit an LOI for the current call, you can look through these sites for other opportunities offered throughout the year.
Because isotope capabilities are not limited just to the FICUS proposals. If you already submitted an LOI in today's discussion, well, then our discussion will hopefully help you how to refine your proposal. And those proposals are due on May 3.
And if you're interested in what you'll hear today but you didn't mention stable isotopes in your LOI, that's not a problem. It's still possible to include them in the full proposal. In a moment, I'm going to turn it over to Mary Lipton, a scientist at EMSL for an overview of technologies there. After Mary, I'm going to provide some information on JGI's metagenomic capabilities.
And then finally, we're going to hear from Jim Moran from Pacific Northwest National Lab on how the capabilities from both facilities can be combined into compelling research projects. Again, I encourage you to submit your questions through the Q&A tool as we go and we'll do our best to answer them. OK, Mary, the stage is yours.
Go ahead. MARY LIPTON: All right. Thanks, Rex. I'm going to share my screen. I'm hoping that people can see this. Today, I'm going to be talking about some of the available isotope probing resources we have at EMSL.
And I'd like to thank you all for taking some time out of your morning or afternoon, depending on where you are, to attend our webinar. EMSL, if you've worked with us before, has changed slightly in the way we have organized in that we're now organized not around capabilities, but around scientific integrated research platforms, so we can more align with the science of the users that are coming in rather than just focus on a specific capability. But today, I'm going to talk about the ways that we've used stable isotope probing in some of the research we do.
So the first one is in our plant and ecosystem phenotyping, where we have a number of capabilities to expose systems to isotopes. For instance, we have growth facilities where we can grow plants or expose soils to isotopically labeled gases in our incubations systems. And so we can look at metabolic and substrate flux through these systems by exposing the gas to the plant or the soil and then tracking that through the system. We also have the ability to track gas production in real time using a real-time isotopic monitoring system that allows us to put a system in a chamber attached to GCMS that allows us to get the monitor a number of gases all at the same time, as well as the isotope logs.
So for instance, if we want to look at a plant system that is undergoing photosynthesis, we can turn the light on, we can turn the light off and look at not just C12, CO2, but C13, CO2, and O16 and O18 off-gassing at exactly the same time. The next thing we're able to do is a quantitative isotope characterization. And in this case, what some of the things we're able to do is metabolic flux analysis using NMR or GC-MS, where we can take, say, a synthetic biology system and look for proteogenic amino acids and other metabolites for synthetic biology. We're also able to do bulk isotope analysis by our isotope-ratio mass spectrometry capabilities. And one of the things we've been doing is taking our systems, at least our soil systems, and segregating the biomass into metabolites, proteins, and lipids, and then using IRMS to figure out where did the isotope go and what sort of molecule did it go into.
So in a bulk way, did the fate of the substrate in a metabolite? Is it in a protein? Is it in a lipid? And we can get those bulk numbers using isotope-ratio mass spectrometry. But sometimes, we want to actually know a little bit more about what type of metabolite it went into or what type of protein it went into. So we can take these samples further and look for biomolecules speciation. So for instance, we can look at the protein fraction from our extraction and do proteomics and then look for the labeled protein within our system. So for instance, you can see an isotopic pattern-- I don't know whether you can see my cursor-- but an isotopic pattern here.
And what we do in the mass spectrometer without upfront separation is just look for a taling that indicates that this is labeled. We can also do this with metabolites and lipids to find out what sort of biomolecules were labeled after a specific experiment. But then we also might want to know not just what sort of species are labeled but where in your system they are labeled. And so we can do spatial determination of where the isotopes went in our system by doing something like nanosense. And so nanosense takes a sample and then it looks for where did the different isotopes go in that sample and they raster across their sample, it's destructive. But you can tell what isotope went where in your sample.
Now if you want to add a little more resolution to that, you can do a mass spec imaging, which also gives you label metabolites-- the identification of labeled metabolites and proteins at different resolutions. And so you can see SIMS right here but we also have MALDI, nanoDESI, lacI, and a number of other types of ionization that have different resolutions. So depending on the resolution of your sample and what you want to know about your isotope, you can use a number of these different methods that let you see spatially where did the isotope go in our system. Now we can also take the isotope-ratio mass spectrometry capabilities and do laser ablation.
And Jim's going to talk a little more about this later, but what this does is the laser actually drills a hole into the system to liberate the mass. And then it goes into the isotope-ratio mass spectrometer. What you can see here is actually a hair, and the laser has drilled into the hair into these little samples. And so you can imagine this being a plant root or a part of a microbial mat, if you want to do layering in the microbial mat, to figure out where did the isotope land after you get it into your system. And then of course, we have high-performance computing, which can take all of this data together to understand the fate and transport through our ecosystems. So the other thing we can do is we can work with samples that are not easily accessible.
And so one of the technologies we have is mass spectrometry and NMR to do organic matter analysis. We specifically do this on soils, where we can extract the organic matter and then we can characterize this organic matter through these systems. Now NMR and mass spectrometry are very sensitive to isotopes. NMR actually looks at the isotopes.
And again, with mass spectrometry, we're looking for that mass shift. And so we can use these capabilities to look at what sort of species did the isotopes go into. And then we're also able to use solid-state NMR to track isotopes into plant and fungal cell walls, and so we can figure out, did the isotope go into the cell wall? We've been doing this with specific plants to look at what types of lignin is being made. And so you can use solid-state NMR in a non-destructive way to look at the speciation of the lignin in your plant cell wall or what happens in your fungal cell wall and where does that isotope go. And so that is a very, very quick overview of the different capabilities that we have at EMSL. I'd like to point out that we do have ways of interacting with the EMSL.
We have EMSL summer school coming up on July 12 through 16, and we have our integration meeting on October 4 through 7. I believe all of these are going to be virtual. We have a YouTube channel that many of these capabilities are explained in a lot more depth than I had time to go through today.
And I would like to point out that both Jim and I are contacts for EMSL if you want to talk about any of these isotope capabilities that I went through very, very quickly-- I'm just realizing. So please put your questions in the Chat and I want to hand this over to Rex to talk about some stable isotope probing in metagenomics. REX MALMSTROM: Thanks, Mary.
All right, so now is a chance to hear a little bit of the capabilities at JGI related to isotopes. We don't have the array. You just saw at EMSL, our main method is stable isotope probing metagenomics. And so I'm going to get a chance to tell you about this in a bit more detail. So this example that I'm going to give happens to be using C13, but I want to let you know that we can do this type of work within N15, and also with oxygen 18.
And before I start, I just want to encourage anyone who's interested in learning more about stable isotope probing metagenomics, check out this link here in the slide. They have some great videos and a great protocol. I'm just going to borrow this one for a second.
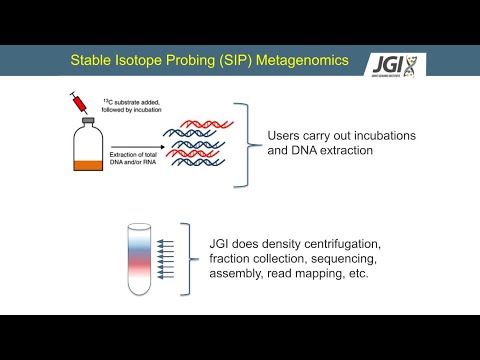
OK, well, any of these type of studies, they start with an incubation where you're going to expose a community to a stable isotope and you're going to grow where you're going to carry that incubation, where you make an addition of an unlabeled substrate in one set and a labeled substrate in the other. And you let these communities grow. And the C13 one in this example, microbes are going to consume that C13, the isotope will get incorporated into their DNA, and that's going to make their DNA a little bit heavier. You can then put that DNA into an ultracentrifuge with the caesium chloride density gradient and separate the DNA out based on its density. Then you can collect these different fractions, separating the heavier DNA from the lighter DNA. Once you've collected those fractions, then you can go on into different work with that DNA.
This cartoon just goes over some of those key points. Again, you're doing an incubation with an isotopically labeled substrate that gets incorporated into the DNA, at least into the organisms using it. And then you centrifuge it and separate out the DNA based on density. So although I just told you, there's an important point to clear up. This idea on the end that there is a heavy and a light fraction of this binary, this is really not true.
This is not true, it's a much more complicated situation. In fact, DNA, how it separates along the gradient is based upon the GC as well. And so instead of tight bands, what we actually have this spread and you could see here, you know, we might have the low GC up here in the less dense, a higher GC over here.
And then when you compare that to what's going on with labeled DNA, there's this overlap between the unlabeled high GC DNA and now the isotopically labeled low GC DNA. So it's a bit more complicated. It's actually blurred together. And so what you have to do is collect many different fractions and then look at all of them.
Here's another way of illustrating that point where, in this case, we're looking at DNA from an isolate. So it's a bit of an idealized scenario. But this is an isolate that's been growing on different levels of label.
And you can see we have zero label there at this lower density, and then as you increase the amount of labeling, they just start shifting down. And if you're imagining doing this in a real experiment, like a metagenome, you kind of have to look at this organism by organism and how does the distribution of their DNA look in an unlabeled control versus a labeled treatment. And so that can start to get a little bit complicated.
So anyway, I just wanted to emphasize that this is not a binary. It's not a heavy-light scenario. It's a bit more complicated. Well, how does this actually work if you want to do metagenomic with JGI? As a user, you would be carrying out the incubations and the DNA extraction.
And then you would send that to JGI and we will do the rest-- the density centrifugation, the fracture collection, sequencing, assembly, read mapping, and so on. That's how it breaks. Yeah, we can't do the labeling growth experiments at JGI. So when we do that, when this material is sent to us and we're able to process it, we use different automation along the process. And this is important, not necessarily because to go really fast, but as you can imagine, it starts to get kind of complicated.
If you have, say, 40 different initial samples of biological replicates and controls, if you have 40 of them and each of those initial samples turns into 12 sequencing libraries, we now have 500 libraries. And it gets tricky to keep track of all of them. So we use this automation and barcoding strategies really to keep track, and it just makes it easier to do these much larger experiments without getting confused and being able to keep track of everything. We also use automated fraction collection, which is much more reliable and reproducible than sort of the traditional way of just collecting drops in a tube and having a graduate student or a technician just sort of switching and different tools for different fractions. That's tedious and time consuming, and that's a really tough thing. So we can use automation to make it a lot easier and you wouldn't have to do it yourself.
It also makes it very easy to change how many fractions you want to do for a particular set of samples. So this helps. Well, after we get those fractions, what do we do with them? I'm going to have a couple of numbers here. It's always a little dicey to throw out numbers because they can change pretty fast.
But more or less, we'll collect something like eight to 12 fractions with DNA, and those will then go on for shotgun sequencing, anywhere from three to five giga base pairs per fraction. These rates then can be combined. We can take all the different fractions and combine them together for an assembly to generate genome bins or bags, and then we can map the reads back to the contigs and MAGs to determine the abundance along the density gradient. So note here, I didn't say anything about 16S. And that's because we don't do 16S sequencing of the fractions, which is the traditional approach with DNA, so we do shotgun sequencing.
However, if you wanted to pair the sort of metagenomics that I'm talking about with traditional 16S screen of the different fractions, you can just ask and we'll send you an aliquot of the fractionated DNA, and that's something you can do on your own. OK, well, so that was a bit of an overview of how the process works at JGI. A little bit about what you get. But I just wanted to finish up with a note about successful metagenomic projects, because they depend entirely on the quality of the incubations.
I mean, the first key is that you need to have enough isotope enrichment of the DNA to see a shift in the density gradient. And again, it's always tough to throw out a number here because these change and also it depends. But very roughly, you're going to need more than 10% atom enrichment in your DNA in order to detect the shift of the phylogenetic group. It's also important to remember in this process that the isotopes are incorporated into DNA when their genome is copied and cells divide. So it's important then to consider the time and how long your incubations go. If, for example, you were working with systems from the deep ocean where doubling times are measured on the order of weeks, it's important then you can't carry out an incubation for three or four days.
The cells just didn't have a chance to divide. So it's not going to get incorporated into the DNA. So it's important to consider this-- the system you're working with, how long it's going to take for the cells to grow, how many divisions they'll go through, and how much isotope they might be able to incorporate.
I know that's a tough thing. We can't really tell you what it's going to be for your system. You guys are the experts in your system. But hopefully, you can have a rough estimate.
And then you can also do time series. And we have the ability at JGI to prescreen the DNA for enrichment before we go through the fractionation of sequencing. And we can do this on a much larger number of samples than we would actually go through the whole process. So it's possible to do this sort of prescreening to triage the samples and identify the ones that will-- or really should go forward with sequencing. OK, so that was the key. That is really the key to any of these isotope studies, it's the quality of the incubations.
All right, that is it for me for the quick overview of metagenomics at JGI. I'm going to turn it over now to Jim Moran, if I can get a hold of my Zoom controls here. They just disappeared on me. There we go.
I can stop sharing my screen now. So I'm going to turn it over to Jim Moran. He's a scientist at Pacific Northwest National Lab, and he's going to tell you a bit more about some of these techniques and how they can be combined together to do really interesting science. JIM MORAN: Great. Thanks very much, Rex. Apologies, I'm a little slow on the Zoom today.
See if I can get this started. So I only have a few slides so we don't have a ton of time to go over this. So what I'm hoping to do with these sites is really provide a few examples of some things that we can do. If you're interested in something that's not listed in these slides, please don't take its exclusion as an indication to mean that we can't do it or something like that. Instead, let's look at the themes that are here and hopefully these can offer something to build upon for questions or future work together. So as we get started, there's a lot of scientific tools out there that can do a lot of different things.
And we all know that one of the greatest challenges is linking the proper tool with a proper scientific question, right? Let's not be limited by tools, but also let's not measure everything we possibly can because we want to stay focused on what the science and how the science is directing us. I want to use rhizosphere systems as one example of this. And the reason for this is it is at least two-fold. One, rhizosphere systems, we know, are important.
They're often referred to as the thin, small, or really spatially constrained region around plant roots. But we know that beyond that, they have global implications. We're going to carbon cycling, plant health, soil health, even hydrologic cycling. Despite all that, there's still a whole host of unknowns about how rhizosphere functions.
And again, I will say that rhizosphere is maybe the epitome in some ways a true biogeochemical ecosystems. It's not only dominated by plants, there are only microbial considerations or only geochemistry or only for water chemistry. But everything is really drawn together, and it's only the interaction of all of those things that enables rhizosphere to function in the way that we, as a society, depend upon and often we, as scientists, are really sort of entranced by and want to figure out and better understand. And it's questions like this, I think, these really challenging questions, that stable isotopes can offer a great tool for unraveling, right? Obviously, isotope studies aren't the answer to everything and we're not going to be able to unravel everything you ever want to know about rhizosphere systems using isotopes. But using some of the tools that I want to just take a few slides to describe now, I think we can eliminate some of the long-standing knowledge gaps that we do tend to encounter.
So in order to demonstrate some of these tools that we have, I'm going to show you some example data that's largely collected from very similar systems. I don't have a lot of field data to discuss. Instead, what we're going to focus on now are largely lab-based systems cultured in rhizoboxes, things like that. Oftentimes, we're using actual soil. In those cases, if the soil is collected from the Kellog Biological Station in Michigan as part of the Great Lakes Bioenergy Research Center-- and any time I'm using a plant, I've been using switchgrass to demonstrate these things. When you think about this box-- I mean, obviously, it's a rhizobox we've brought in the lab to simplify things.
But it's still not a simple system whatsoever at all. You know, despite the fact that you literally can pick it up in your hand, it still retains some of the complexity that you would expect in an actual field system. And we're going to try to highlight that a little bit. As you think about that system, that rhizobox, we often stumble across some of these what would initially appear to be relatively simple questions, right? Who is active? How much of this process is happening? Where is it happening? You know, rhizosphere, being a very spatially constrained system. What processes are being used? Or how are the reactions that were observing at the higher range, how are they actually happening from both the geochemical or even biochemical pathway? And then also, what's the timing? Or when does that happen? And I'm asking these questions-- who, how much, where, how, when-- often we're asking them, in terms of specific components. So when our plant inputs to good oxidation or other processes, when are they applied to soil? And where do they go? You know, similar questions about the microbial contributions.
But maybe also, who is active? When are they active? What are they doing? Similar types of approaches can help solve or could be involved with a better understanding nutrient exchange, or just how poor water and some of the geochemistry there is taking place, and when. So what I'm going to attempt to do is highlight these types of questions, who, how much, where, how, and when using a rhizosphere system as an example. And I want to go through some of the techniques that are available at EMSL and JGI and how they might help to illuminate answers to those questions. And again, this is only a description of a partial subset of the capabilities that could be accessible to you as an EMSL and JGI user. But let's get started.
So the first question here is, how much? As you begin to get involved with isotope studies, you realize that "how much" is quite an open-ended question. You can address that with multiple levels of precision. And really, the science has to guide how much precision you need in making that assessment. But that being said, for stable isotope analysis of light elements like carbon, hydrogen, nitrogen, oxygen, really, the ideal way to quantify and answer that "how much" question is typically with isotope-ratio mass spectrometry. This is the workhorse of the traditional stable isotope fields.
It really helps quantify-- quantify is basically what it does, right? That what it's great at. What it can do is quantify how much stable isotope is in whatever sample you give it. So if you literally take a spoon and carve out some soil, it can tell you how much 13C, 15N, or whatever tracer or natural abundance element might be in that particular sample. You can get a little bit tricky. You can sort of break down the sample into different components and then get a little bit more resolution in that quantification, right? So again, you can pull the full soil, you could take out specific roots.
And since you dig out specific roots, you have a tissue-specific approach, you can also look at specific phases. So if you're interested in just dissolved organic carbon, you could pull out the DOC from a sample and get a very good quantification of how much tracer is in that DOC or the CO2 released. Or you could use some chemical separations and look instead at specific amino acids or different lipids that might be linked to specific biomarkers to look at pigments. All of these are just a subset of the things that you can look at.
But again, think about it, we're asking really that fundamental question of "how much." You know, you apply the tracer, how much that tracer is going into these different bins? You have an example here on the right side of this slide. This is showing a split root rhizobox here. We have a switchgrass plant and we're trying to answer the question of, is there spatial heterogeneity in root exudation processes linked to the availability of the nutrients? So we have phosphorus availability on one side of this rhizobox and not on the other, but that's the only difference between those two halves. And we've applied a 13CO2 tracer to the plant and we're just using isotope-ratio mass spectrometry to quantify any differences in the amount of root exudate investment that the plant is making on both sides of this box.
So again, IRMS really helping us to answer in most cases, this question of "how much." But as Mary alluded to, we can also sometimes link that to a spatial comparison, right? This is something that we developed in our lab here. I mean, it is gaining a larger popularity. It's still somewhat of a niche approach, but there are some other labs that are doing as well. Here, as Mary mentioned, you've already seen this picture. Here you can see a hair that has three different holes drilled in it with a laser.
Each of those holes are about 50 microns in diameter. So it gives you a sense for the spatial resolution that we can attain when you have a fairly high carbon substrate here. But each of those holes also have plenty of material for making multiple 13C measurements.
We get really good precision on those measurements, so we can begin to answer questions that might link quantification with a spatial analysis, right? This is another hair here. You can just see it. It's kind of neat, you can really blast that hair with the laser and you saw it carve out of those little particulates. And those particulates we then combust and send the resulting combustion CO2 to the IRMS for making isotope measurements. You can take that spatial approach and you can apply it to a plant system. So here we have, on the left, an image, a visual image, some switchgrass roots growing in soil.
And then on the right, each one of these marks is where we've done one of those spatially prescribed laser ablations and measured the exact amount of 13C in each of those spots. And what you can see is that this main root has a lot of 13C in it, you can see by that deeper red color. And that's all coming from recent photosynthate, where we've applied a 13C tracer to the plant. But what you can also begin to see is a lot of variability in how much of that recent photosynthate that 13C traces is getting into different roots.
This root B, for instance, is seeing a lot more of carbon allocation than even roots C or D in this case. We don't have to look just at roots. We can also begin to quantify root exudation. So this is a slightly different image that you can see here. But you can see these pits. Soil isn't quite as clean as here when it comes to ablation.
You can still see these darker pits. That's where we've taken of laser ablation sample. Each one of these, we use a 50 micron spot size in the laser but these spots get a little bit bigger in soil.
But we still have a fair degree of spatial resolution. And what you can see is when you label the plant, we can see this tailing off of root oxidation as you get further away from the surface of the root. So by about somewhere between the order of 1, 1 and 1/2 millimeters from the root, we've really seen a tailing off of that root exudation back to almost background levels of the organic carbon in the soil. So that was a really great quantification and some spatial analysis.
Mary also mentioned NanoSIMS. NanoSIMS can also do some very good quantification. Not quite as good as IRMS, but NanoSIMS has spectacular spatial resolution. As its name might imply, really you can get on the order of tens of nanometers spatial resolution. You can also look at more than one isotope simultaneously.
They have to be somewhat closely paired on the periodic table. You can't look at isotopes at the top and the bottom at the same time, but still, in this example, you can see that this is a switchgrass root. And you can see that we're looking at uptake of 15N. I believe we applied it as nitrate in this one.
And then also 13C in that root. Also from a 13 CO2 tracer. So great, great spatial resolution.
One thing to keep in mind is the spatial resolution is incredible, but it also comes at the expense of how much of the surface area of that plant you can analyze. You're a little bit constrained. If you're looking to really understand what's happening on the almost the cellular level or something very close in, NanoSIMS is a great tool. If you're looking for trends that are occurring over millimeters or centimeters, you might not want to consider NanoSIMS quite as extensively because it's hard to scan over those spatial regions. So moving on a little bit, to answer this "who" question.
Rex provided an overview. You've already seen this slide. You know that we can add a 13C C-labeled substrate, for instance, and try to physically change the behavior of the resulting DNA.
You're focusing largely on density separation, with obviously, this example here, being a mostly unrealistic, especially in a soil system. But we can think also a little bit-- I mean, Rex mentioned some of the different labeling approaches that you can use with DNA shift. Certainly, each of these different labels can help you address a slightly different question while really, they're all under the umbrella of trying to identify who. You know, who is active under these specific conditions? That might be something that an 18O tracer can use. Who is active at consuming a specific substrate? You might have a 13C labelled substrate.
Or, you know, who is participating in different components of the nitrogen cycle? You might have a 15N labeled substrate addition in that case as well. Again, all really trying to get at this question of who. There's some other options for getting at that question of who as well, and obviously, there's trade offs in all of them. We can also turn to proteomics.
We get some resolution of who somewhat constrained by depth of metagenomes and things of that nature. But we can also begin-- what we're beginning to do is to provide some level of understanding of where in that question in the proteomics analysis as well. And to some degree, you can also use proteomics to hopefully begin assembling metabolic pathways and potentially have some insights into this how things are proceeding as well. So I mentioned, we're trying to make some improvements in the proteomic pipeline. Obviously, you can do a tremendous amount with traditional proteomics.
What we're working in EMSL is to try to improve our sensitivity of analysis basically to enable smaller sample sizes. And when you can have less sample size, you have the opportunity to really spatially focus how you collect that sample and then get some-- the proteomics data, in that case, we would be able to interpret that in some sort of spatial context. As Mary mentioned, we're also making some advancements in trying to use a 13C and 15N tracer coupled with proteomics to better understand things like taxonomic activity, substrate utilization, things of that nature. And this is being pursued by other publications as well, other labs besides just EMSL but we're pretty excited by some of the results that we do have.
So I want to take you through one example we have. We're going to go back to our rhizobox. It's been a great way to demonstrate some of our capabilities. When you think about spatial resolve proteomics, there's a couple of things that we're doing.
One, we can really apply this sort of blotting technique where we put a membrane on the surface of the soil, remove those mobile proteins, stick them onto the membrane, and then we can do a couple of things. We can map activities. We've demonstrated mapping phosphatase activities.
And then you can also map the protein distribution. And on that protein distribution map, you can then go in and spacially subsample that membrane and perform proteomics on what you've pulled off that membrane to enable some sort of spatial resolution. I'm going to give a little bit more of an example on that. Here is an actual example instead of a schematic. You can see here a switchgrass root system.
We've done this membrane blotting, and we can map phosphatase activity and also total protein distribution. At some amount of spatial scale, we see that when we've applied some organic resources-- this is an organic powdered plant material that we've pelleted and put in the rooting zone of soot transplant-- we can also see that root 1 expresses a little bit more phosphatase activity in close proximity to it to root 1 versus root 2, which is next to our control. Potentially some indication that what we're seeing in a spatial context is in agreement with the experiment that we've done here. But I want to push a little bit further. Here's another example. You can see these membranes-- We're doing this membrane blotting sort of extraction.
Here's a particular root that we were targeting. Apply this membrane. And then we can look at the total protein distribution. And what you see in this final panel on the right is you see that same membrane with some holes taken out of it. So we've punched those holes out, and what we're doing is performing proteomic analysis on what proteins are attached to the membrane that is physically correlatable back to different locations on the host plant root.
When you put all this together, we can begin to ask questions like, can we distinguish the proteome at the root tip from the mid-root? Or more mature rhizosphere up from the root tip? And certainly, we can dig into this more offline. But certainly, we're seeing that. We're seeing a real shift in the microbial in-plant community structure that we're observing with proteomics.
And obviously, we want to pursue that a little bit from a functional component as well. We can also apply it, as Mary mentioned, a 13C tracer. So back to this rhizobox that we saw before, we labeled this plant with a 13CO2 tracer. And then we ask the question, OK, can we specifically pull out proteins that show incorporation of this 13C tracer? Can we do that on both sides of those rhizoboxes where we have different sort of ecosystem dynamics brought about upon by the abundance or lack of added phosphorus? And what we're seeing, first of all, is that we do see a number of proteins. Not tons, but we see a number of proteins that show incorporation of that, that 13C label.
And we're also seeing some differences between that in the two phosphorus treatments that we observed. So for instance, we have no phosphorus addition. We're seeing much less influence from the switchgrass plant, many fewer proteins related to switchgrass than we are in the case we're seeing added phosphorus. And I will also mention that this is in contrast to the overall root exudation patterns, which don't seem to show much distinction between the two conditions. But hopefully, this is a reasonable example of how we can look for things that are specifically active using that, in this case, 13C tracer. So last example here because we're running out of time.
But Mary mentioned this real-time aspect capability. It's a chance to look at headspaces over soil or headspaces around plants, and in real time, provide some sort of temporal assessment. You know, what organic compounds containing a label or nitrogen label are being produced by the system at a given amount of time. We're adapting this a little bit so we have a system similar to this.
Again, back to our rhizobox system. We have a system that we're looking forward to testing where we apply a 13C tracer to the leaf portion of a plant, but we exclude that 13C tracer from a soil headspace chamber. So what we're doing, we have some initial data demonstrating that we can show uptake of 13CO2 by the plant transport of that result in organic carbon into the soil, release of that carbon into the soil. And then remineralization to CO2 by a variety of either a combination of plant and microbial processes.
But this can all be tracked in time, right? So we can begin to ask questions of when. Is there diurnal cycling? Does the timing differ when you apply a specific stress or perturbation to the system? And potentially linking that also to light volatile organics that are coming off, how can we manipulate this system and then observe that resulting change in the larger ecosystem by focusing only on that headspaces above the soil? So I realize we've gone through quite a bit. Again, this was not meant to be an exhaustive description of some of the capabilities at EMSL and JGI.
There's certainly more to offer. But hopefully, this has gotten your interest a little bit. And please reach out to me, Mary, or Rex with any sort of questions that you might have because we always really enjoy chatting about some of the things that we can do.
So thanks very much and back to you, Rex. REX MALMSTROM: Thank you, Jim. I think that was a great presentation of some of the different capabilities of the two facilities and how they can be combined for some pretty interesting the questions. We have a few minutes to answer questions that are coming in now. So Jim and Mary, if you guys want to, you can turn you cameras on and unmute yourself, and I'll start with some of these questions here.
I think we'll start with a question for you, Jim. There's a question about the field applications of rhizoboxes, saying-- I'm just going to read this here. They're interested in tracing isotopes through spatially separated groups in an ecosystem, like a biological soil crust.
I think they're just asking for a little bit more information about how that might work with a rhizobox or if it could work with the rhizobox. Oh, you're muted here. JIM MORAN: Sorry about that. Yeah. Certainly, I guess the rhizobox might not be the critical component. I think it would be a great thing that this could be done in a rhizobox but also in a field system.
Really, the hardest thing-- well, there's two-fold. One, we need to get that 13C tracer in, if you're looking to use the tracer to do the spatial analysis. And then the other real challenging thing is harvesting the sample in a way that you're not disrupting the spatial organization.
So as long as we can harvest the sample and then have a sample, you know, half-inch round diameter or something like that, sort of soil puck that captures that two-dimensional spatial organization, then we can certainly put that in the laser system and provide spatial analysis of that. Hopefully, that addresses the question. But let me know.
We can talk more offline as well. REX MALMSTROM: OK, thanks Jim. There were a couple of questions that came in related to DNA-SIP and I'll just handle a few of those really quickly. So there was a question asking how much DNA is required to do this. And the answer is, the more the better.
We want to put it at the very least microgram of DNA into anyone's centrifuge tube. So that means we need, at the very least, 2 micrograms to start with just in case something goes wrong. So the answer is, you need to be able to extract micrograms worth of DNA for any one individual sample. Another question about how we do the prescreening of DNA at JGI to try and measure the isotope enrichment. That's done through the LCMS. We do a hybridization of the DNA and we look at some of the bases that comes out.
We just see how at risk they are and estimate the overall atom enrichment within the bulk community DNA. And that gives us a rough estimate of, like, if it's worthwhile even pursuing. And then finally, what's the turnaround time? And that's a good question. I think, like a lot of folks, we are impacted by COVID and many other different things and how much we can go through. It takes a few months to do this from start to finish, to do all the QCs, the separation, library sequencing, and assembly. It's going to take a few months.
OK, let's see. A question here. I'm going to go for Mary. Can radioactive isotopes be used? MARY LIPTON: No, we cannot take radioactive isotopes into EMSL and use them on the instruments because it has a long-term negative effect on utilization of those instruments.
So at the moment, we are not able to do anything radioactive with the technologies that I've talked about. REX MALMSTROM: OK. I have another question for you, Mary. Saying, how well suited is the LA IRMS for tracking N15? The example was with C13.
Can you do it with N15? MARY LIPTON: Yes. And Jim can answer this question as well, but yes, we can look at N15 as well as C13 as well as other isotopes using the IRMS. It's just a matter of what you're tuned to be looking for. JIM MORAN: Yes. So we can do that. Right now, we haven't however worked out the ways to do that with the laser ablation IRMS so we don't quite have the spatial resolution at tens fo microns or 100 microns, whatever you want to call it.
But NanoSIMS can do that at a really fine scale, and we can also easily measure the 15N at the bulk level, if that helps at all. And we're working on trying to develop a better sample handling techniques to do that 15N because a lot of people are interested in it. But we haven't got that quite figured out yet. REX MALMSTROM: OK, thank you, Jim.
Jim, I have a question for you here regarding the real-time trace gas monitoring. Can this be conducted while you're doing a 13 CO2 plant labeling experiment? And if so, how do you resolve the respiration cell respiration? JIM MORAN: Yeah, great question, right? The way we've accomplished this so far is that we physically separate the 13 CO2 that we're using to label the leaves from the soil. So we have a separate sort of air chamber around the leaves and then an air gap, regular atmosphere, we've done our absolute best to really seal that gap. It's kind of hard around some stems of the live plant but we have an approach that we think that works.
And then we have that second sort of chamber above the soil headspace. Right now, I would say that we don't have a great way to resolve root respiration from microbial processes. But I can say, at least, we're not mixing the tracer with the CO2 produced at the end.
It's only a partial answer, I don't know if, Mary, you have any other thoughts to add to that. MARY LIPTON: No, I think you did a great job of saying, yes, if we can somehow isolate the root from the CO2 that we're putting in there-- and we have thought about this and are attempting it-- we can measure the respiration. One of the things we've been trying is to actually try to put a probe down into the soil to get the respiration of the soil directly without actually trying to sample the gas that's above the soil. REX MALMSTROM: OK.
Jennifer, I hope that answered your question. So here is a question related to DNA-SIP. It also then becomes more general. And it's asking, are there any special considerations for working with N15? Does the label need to be relatively stronger than C13? The answer is yes. Certainly, for DNA-SIP just because there are fewer nitrogen atoms than there are carbon atoms in DNA. So you need to get a little bit more labeling going on with nitrogen to see that separation.
I'm afraid I can't give you a specific number of what that needs to be, but I mean, you can just sort of estimate from the relative amount of carbon and nitrogen and still try to target this idea of wanting to have greater than 10% atom enrichment in your target organism in order to separate them by DNA-SIP. But as DNA-SIP, Mary, when it comes to the various isotope capabilities there, how much labeling is needed? MARY LIPTON: So it depends on the capabilities. If we're talking about speciation, protein-SIP, lipids, metabolites, actually, it gets to the point where sometimes more label can be detrimental if you got up to too much, because it shifts your masses. And since we're using a mass spectrometer to look at those masses, it makes it hard to identify the biomolecules in the mass spectrometer. If we're using NMR, the opposite is true. The more the label we have, the better we can see it.
So it's really analytic dependent. I would say, we probably don't need more N15 than we need than we need C13 to actually see it. But as you said, there's still fewer nitrogen and a lot of our biomolecules than there are carbons. So the bottom line is it's analytic dependent. And if you have a specific question about one of the things we do, I would ask about the specific analytic. REX MALMSTROM: OK.
Question here for Jim. Since you were able to identify the major proteins that incorporated C13 and sort of the isotope of proteomics, what about doing this with metabolites? JIM MORAN: Yeah, I might hand this one to Mary in part. And certainly, I think, well, let me Mary answer. Go ahead, Mary. MARY LIPTON: So with the way we did this, we transferred the proteins to the substrate, the paper, to get them off. For as much as we can get the metabolites on the paper, we can do that.
But I think the better approach would be to further develop our imaging capabilities to do it directly with some aspect imaging. A lot of people use MALDI imaging, but then you have to get the MALDI matrix into the soil and mix it up. So I think there's some other imaging technologies that we could use to target the metabolites. So I don't know that I would take the exact same approach that we do with proteins but I think that we have a way of doing this with metabolites.
JIM MORAN: The answer to the question might also hinge on if you're interested in a specific metabolite. If you could do some sort of targeted approach and you needed a lot of quantification, you could potentially link that with IRMS. Or if you're interested in a more global approach, I think some of the techniques that Mary mentioned are probably the place to start. REX MALMSTROM: There was a question about analysis in KBase.
DOE KBase is a data analysis platform, really fantastic for doing this type of work. Certainly, for any type of DNA sequencing work and combining these other types of omics. The quick answer is, currently, metagenomic, there are not existing narratives but they're definitely-- they're on the to do list, to build a road map, to be able to incorporate submetagenomic data sets into KBase. OK, another question.
Let's see. Yes, so can JGI work with labeled RNA? The answer is not right now. That's also on our radar and so hopefully, that will be something we can roll out in the not too distant future. Let's see. We have a new question here. This is going to be tricky to answer, but I'm going to throw it out there anyway.
So how long do plants need to be placed in a 13 CO2 chamber for sufficient labeling? MARY LIPTON: So let me take this one and then I'll pass it to you. Because the question is, are you looking for the label in the plant? Or are you looking for the label in the soil in the rhizosphere? Because obviously, we're looking at a label in a plant. You know, it could be as short as 24 hours. And we actually see uptake of that label into plant material. If you're looking at the deposition into the rhizosphere, so the label has to go into the plant, out through the roots, into the soil, and then picked up by the rhizosphere, then that takes a little bit longer. But Jim, you've done these experiments and you can say how long it took you to get it into the roots.
JIM MORAN: Yeah, so unfortunately, I haven't done as much work as I'd like to with the timing on this. And we do know that some of the work we've done, it takes at least four or five hours before you see a 13 CO2 applied to a plant leaves to get through and respired. But it takes a lot longer to build up 13C level biomass, right? So most of what we've done has really been on the 24 to 48 hour 13C labeling exposure time. But it really brings up a big question as well. You know, how much CO2 do you use? How much are you wanting to perturb the system to get more lable or more tracer in there? And things like that. So I think a lot of these questions are in some ways sort of experiment dependent.
I think what Mary was saying as well is that if you have a plant that's in a very active growth phase, it would take potentially less time than a plant that's beginning to go through senescence or something like that. Something else that we can do is-- what we can do, I think Rex sort of mentioned this on his end as well, that if you do do a time series and you want us to analyze at sort of the highest level, is there enough there that might be something that is a lot easier to run through an initial time series experiment and really help us dial in what might be the right labeling duration for your specific application. REX MALMSTROM: Thank you, Mary.
Thank you, Jim. Great answers. Well, I'm looking at the clock here and we are just about out of time. I think we got through all the submitted questions, but if you think of new questions, please don't hesitate to contact us with us because we'll do our best to answer them. So to Mary, I'm just going to close this out here with a few more slides. So here's some more information to find.
Contact information. And there's a lot of useful information there. So thank you.
Check it out. Thank you for participating in today's events, for those of you who tuned in. And Thanks to Mary and Jim for their talks. And I hope that we were able to answer the questions. If you guys feel that you need more information, please contact us. So my email's here, as well as Mary and Jim's.
You can also reach out to anybody you know at JGI or EMSL. We can always try and address those questions. If you want to learn more about the FICUS program-- where you can request access to these capabilities of both user facilities, you can find more at either of these two sites up here, or again, contact us. You can also keep in touch with the latest, with what's going on at JGI through various social media platforms. You can visit our YouTube channel.
And we also have some podcasts which you might be interested in checking out. All right, that does it for today. Best of luck to all of you with your proposals and your proposal submissions.
We look forward to reading them when they come in. That's it. That's all for now. Take care. Goodbye.
2021-04-09 05:19


